Occurrence and Molecular Characterization of Some Parasitic Copepods (Siphonostomatoida: Pandaridae) on Pelagic Sharks in the Mediterranean Sea
- 1Department of Integrative Marine Ecology, Stazione Zoologica Anton Dohrn, Naples, Italy
- 2Museo Civico di Storia Naturale, Comiso, Italy
- 3Wilderness Studi Ambientali, Palermo, Italy
Copepods of the family Pandaridae are typically ectoparasites of elasmobranch fishes. They display a cosmopolitan distribution and limited host specificity. Published literature on their occurrence on pelagic sharks in the Mediterranean is scarce, often from the past century, or scattered through fish parasite surveys. Moreover, of the 64 valid pandarid species known at present, molecular data from GenBank exists for only 10 species and there are no data from the Mediterranean. In this study, we begin addressing this knowledge gap by exploring the molecular features of some pandarid copepods (i.e., Dinemoura latifolia, Echthrogaleus coleoptratus, Pandarus satyrus, and Phyllothyreus cornutus) and their phylogenetic relationships using new material from pelagic sharks (i.e., Prionace glauca, Isurus oxyrinchus, and Carcharodon carcharias) in the Mediterranean. Genetic distances analysis showed intraspecific variation in the mitochondrial DNA cytochrome oxidase c subunit 1 (mtDNA cox1) sequences and interspecific variations of 0.001–0.081 and 0.196–0.288, respectively, for the small subunit ribosomal DNA (SSU rDNA) and the cox1 gene locus. Phylogenetic analyses of pandarid copepods based on sequences available in GenBank plus the sequences generated by our study revealed two major clades: the first, with strong nodal support, included species of Pandarus, Phyllothyreus, Pannosus, and Pseudopandarus; the second, with weaker nodal support, included species of Achtheinus, Perissopus, Echtrogaleus, Nesippus, and Dinemoura. As most pandarid species are missing from the present analyses, we discuss the limitations of our phylogenetic results. Nevertheless, this study represents a first step toward to yielding new information about the phylogeny of parasitic copepods on pelagic sharks in the Mediterranean.
Introduction
The Order Siphonostomatoida Thorell, 1859 includes 39 families of copepods and encompasses about 75% of all parasitic copepods on fishes (Gunn and Pitt, 2012). Members of the family Pandaridae Edwards, 1840, are typically parasites of external surfaces of elasmobranchs (Kabata, 1979; Izawa, 2010; Bernot and Boxshall, 2017). Pandaridae consists of 23 valid genera with at least 64 recognized species (Walter and Boxshall, 2021). Pandarid life cycles have been not elucidated, however, a life cycle similar to that of Caligidae Burmeister, 1835 has been proposed based on the close phylogenetic relationships between these taxa (Huys et al., 2007; Dippenaar, 2009). The supposed life cycle of Pandaridae includes two free-living nauplius stages, one infective copepodid stage, four parasitic chalimus stages, two parasitic preadult stages, and the parasitic adult stage (Wilson, 1907).
Pandarid copepods are characterized by attachment organs named adhesion pads (Kabata, 1988). Their adhesive surface is formed by a thick cushion of skin with a ridged outer layer (Wilson, 1907). The site of attachment on the host varies depending on tissue tropism and the fundamental niche of the parasite species; they can colonize fins, gills, the cloacal aperture, the mouth, or nasal passages (Benz, 1981, 1986; Rokicki and Bychawska, 1991). Pandarid species are cosmopolitan in their distribution, occurring in warm and temperate waters with most of the species capable of parasitizing more host species (Alvarez and Winfield, 2001).
Published literature on the occurrence of pandarid copepods on pelagic sharks in the Mediterranean is scarce, often from the past century, or scattered through fish parasite surveys (Brian, 1906; Öktener and Trilles, 2009; Öktener et al., 2020). According to the most recent studies, members of Siphonostomatoida remain largely unexplored in terms of their molecular characterization and phylogenetic relationships (Dippenaar, 2009; Bernot et al., 2021). In particular, of the 64 valid pandarid species listed at present, molecular data from GenBank exists for only 10 species and there are no data from the Mediterranean. The present study aimed to report the occurrence (and characterize using a molecular approach) of pandarid copepods obtained opportunistically on shark species off the coast of Sicily (Italy) and to provide newly generated molecular and phylogenetic data to improve knowledge of the poorly known Pandaridae parasites infecting sharks.
Materials and Methods
Sampling and Parasitological Analysis
The material here studied comprised undetermined copepod parasites collected by two co-authors (GI and BZ) under the framework of a project of the Museo Civico di Storia Naturale (MSNC) in Comiso on non-native and rare marine species of the Mediterranean Sea (see Katsanevakis et al., 2020; Deidun et al., 2021). The MSNC is a scientific institution registered at the CITES Secretariat, D.M. 23.03.1994 (Cod. IT030), authorized to take, keep, use and display dead endangered fauna.
The present material encompassed copepod parasites collected from 2003 to 2021 from the coast of Sicily on six pelagic sharks [i.e., three blue sharks, Prionace glauca (Linnaeus, 1758), two shortfin mako sharks, Isurus oxyrinchus Rafinesque, 1810 and one great white shark, Carcharodon carcharias (Linnaeus, 1758)]. The blue sharks were from strandings; the shortfin mako sharks and the great white shark were caught as bycatch (Table 1).
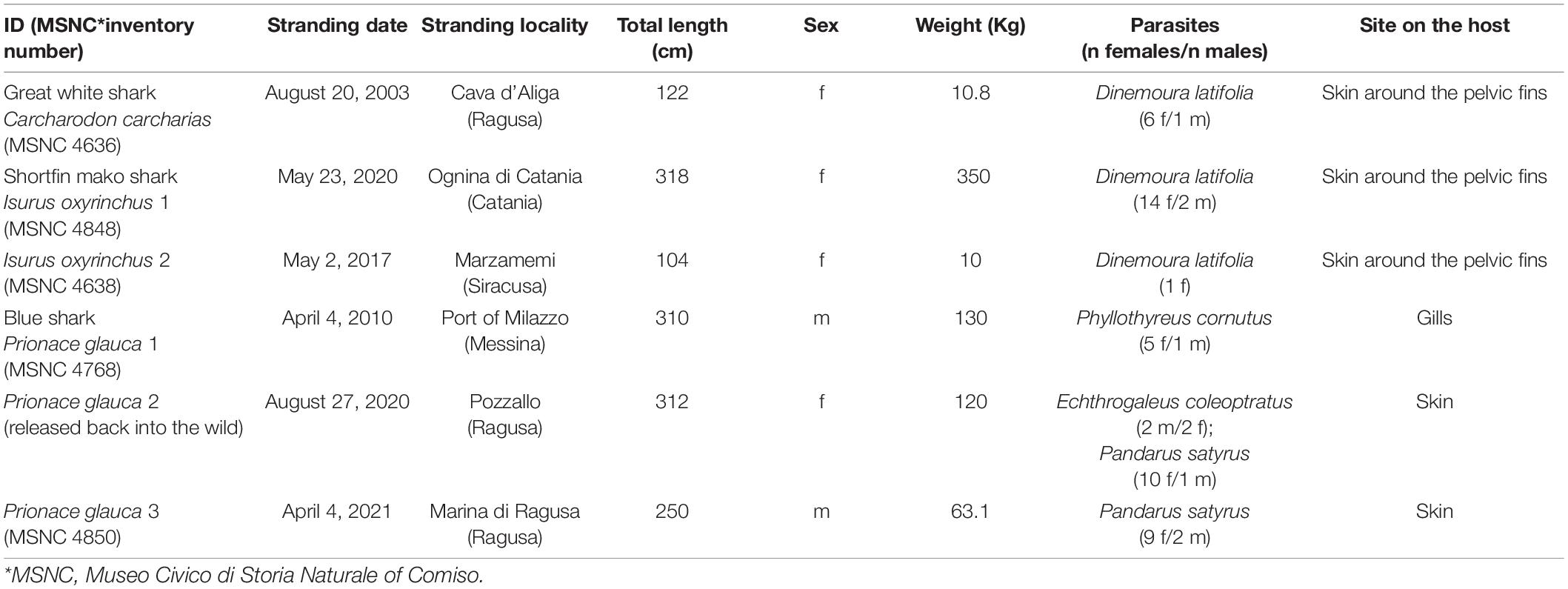
Table 1. Available data of shark individuals examined for pandarid copepods from the coast of Sicily.
The taxonomic identification of sharks followed Compagno (1984). The fishes were weighed, measured (total length) to the nearest 0.1 cm and sexed by visual observation of external characteristics. Copepods from the skin were carefully removed using forceps while gills were removed from carcasses and examined for copepods in Petri dishes under a stereomicroscope. Copepod parasites were counted, washed in physiological saline, and preserved in 70% ethanol (Santoro et al., 2014, 2020). For identification, copepods were sent to the Stazione Zoologica Anton Dohrn in Naples where they were studied using a stereomicroscope and an optical microscope both equipped with the ZEN 3.1 imaging system (Zeiss). Morphological identification of copepods followed the identification keys of Lewis (1966) and Cressey (1967, 1968). After examination, the sharks were prepared and incorporated into the museum collections of the MSNC under inventory numbers as listed in Table 1, except the blue shark #2 which was a live individual rescued, rehabilitated, and released back into the wild after the external examination.
Molecular and Phylogenetic Analyses
Following the morphological identification, genomic DNA was extracted from the antennae of six specimens of D. latifolia, collected from a shortfin mako shark (n = 3) and a great white shark (n = 3), and two specimens of Echthrogaleus coleoptratus (Guérin-Méneville, 1837), three specimens of Pandarus satyrus Leach, 1816 and two specimens of Phyllothyreus cornutus (Milne Edwards, 1840), collected from the blue shark. Genomic DNA extraction was performed using a Quick-gDNA Miniprep Kit (ZYMO RESEARCH), following the manufacturer-recommended protocols, with modification of the incubation period with proteinase K to 3 h.
The small subunit ribosomal DNA (SSU rDNA) (∼1,795 bp) was amplified using the primers 18Sf (5′-TACCTGGTTGATCCTGCCAG-3′) and 18Sr (5′-TAATGA TCCTTCCGCAGGTTCAC-3′) (Huys et al., 2007). The partial sequence of the mitochondrial cytochrome c oxidase subunit 1 (mtDNA cox1) (∼600 bp) was amplified using the primers LCO1490 (5′-GGTCAACAAATCATAAAGATATTGG-3′) and HCO2198 (5′-TAAACTTCAGGGTGACCAAAAAATCA-3′) (Folmer et al., 1994). Both polymerase chain reactions (PCRs) were performed in a 25 μL volume containing 0.6 μL of each primer 10 μM, 2 μL of MgCl2 25 mM (Promega), 5 μL of 5 × buffer (Promega), 0.6 μL of dNTPs 10 mM (Promega), 0.2 μL of Go-Taq Polymerase (5 U/μL) (Promega) and 2 μL of total DNA. PCR temperature conditions for the SSU rDNA were the following: 94°C for 5 min (initial denaturation), followed by 35 cycles at 94°C for 30 s (denaturation), 57°C for 30 s (annealing), 72°C for 30 s (extension) and followed by post-amplification at 72°C for 5 min. PCR cycling parameters for the mtDNA cox1 amplifications were: 95°C for 5 min (initial denaturation), followed by 40 cycles at 95°C for 1 min (denaturation), 45°C for 1 min (annealing), 72°C for 1 min (extension) and followed by post-amplification at 72°C for 7 min. PCR amplicons were purified using the AMPure XP kit (Beckman coulter) following the standard manufacturer-recommended protocol and Sanger sequenced from both strands, with the same primers, through an Automated Capillary Electrophoresis Sequencer 3730 DNA Analyzer (Applied Biosystems), using the BigDye® Terminator v3.1 Cycle Sequencing Kit (Life Technologies).
Contiguous sequences were assembled and edited using MEGAX v. 11 (Kumar et al., 2018). Sequence identity was checked using the Nucleotide Basic Local Alignment Search Tool (BLASTn) (Morgulis et al., 2008). The SSU and cox1 data sets were, respectively, aligned with all sequences of Pandaridae available in GenBank (see Table 2), using ClustalX v. 2.1 (Larkin et al., 2007). Sequences of both genes (SSU + cox1) were concatenated using SequenceMatrix (Vaidya et al., 2011), while the best partition schemes and best-fit models of substitution were identified using Partition Finder (Lanfear et al., 2012) with the Akaike information criterion (AIC; Akaike, 1973). The analyses were performed using the GTR + invgamma substitution model.
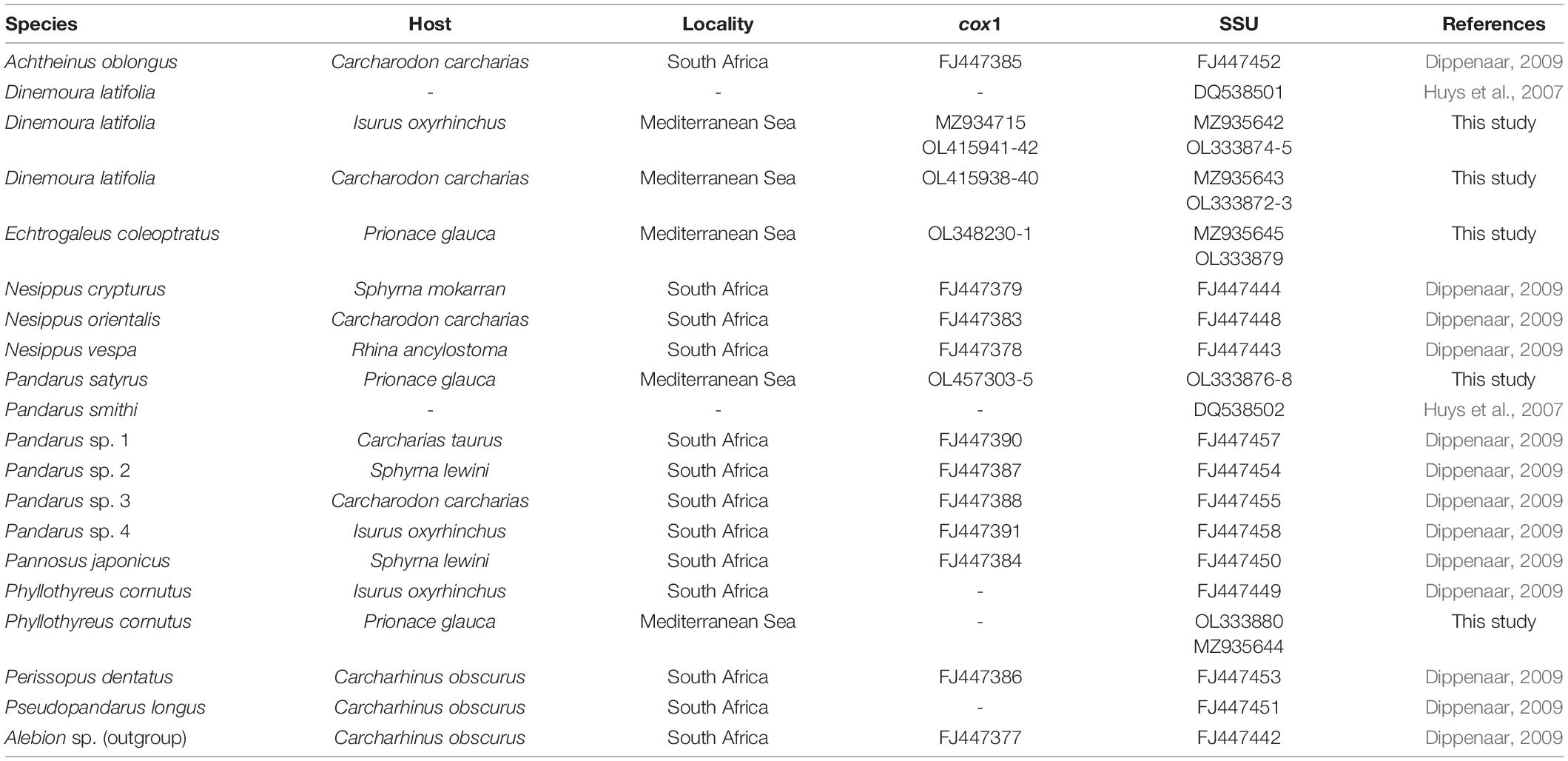
Table 2. Species, host, locality, and accession numbers of cox1 and SSU sequences of pandarid copepods included in the phylogenetic analysis shown in Figure 2.
Sequences obtained in the present study were deposited in GenBank under the accession numbers as listed in Table 2. Genetic distances were computed using the Kimura 2-Parameters (K2P) model (Kimura, 1980) with 1,000 bootstrap re-samplings, by MEGA Software, version 7.0.
The phylogenetic trees were constructed using the maximum likelihood (ML) method by IQ-TREE (Nguyen et al., 2015) with 1,000 ultrafast bootstrap replicates (BS). Clades were considered to have high nodal support if the ML bootstrap resampling ≥ 70%. Due to the phylogenetic position of Pandaridae (see Dippenaar, 2009), the phylogenetic trees were rooted using Alebion Krøyer, 1863, as outgroup. The SSU and cox1 sequences from GenBank included in the phylogenetic trees are listed in Table 2. To corroborate the taxonomic assessment obtained according to the phylogenetic species concept, the species delimitation method on cox1 gene locus was also performed using the assemble species by automatic partitioning (ASAP) (Puillandre et al., 2020).
Results
General Data
Available data from shark individuals examined for pandarid copepods, and species, number, and sex of pandarid copepods found are listed in Table 1. A total of four species of pandarid copepods were morphologically identified. A single species (i.e., D. latifolia) (Figures 1A,B) was found on the skin surfaces of the shortfin mako shark and great white shark, and three species (i.e., E. coleoptratus, P. satyrus and Ph. cornutus) (Figures 1C–H) were collected on the skin and gills of the blue shark (Table 1). Voucher specimens have been deposited in the collection of the Stazione Zoologica Anton Dohrn in Naples with the following accession numbers: SZN-CRUOO2A-2B (D. latifolia), SZN-CRU003A-3B (P. satyrus), SZN-CRUOO4A-4B (Ph. cornutus) and SZN-CRU005A-5B (E. coleoptratus).
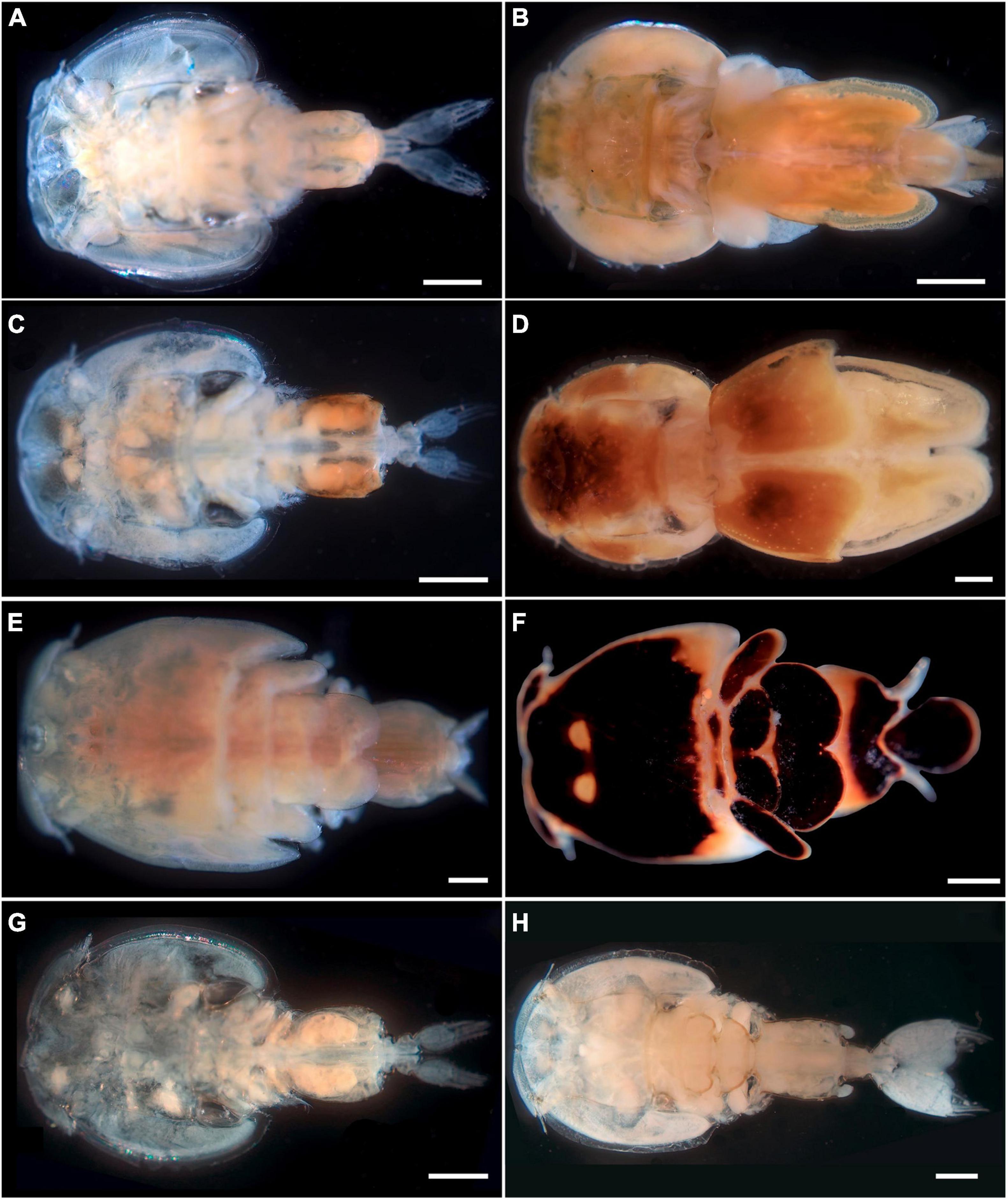
Figure 1. Specimens of pandarid copepods sequenced in the present study. Dinemoura latifolia dorsal view of male (A) (bar scale: 1,000 μm) and female (GenBank: OL415938 and MZ935643) (B) (bar scale: 2,000 μm) from the great white shark; Echtrogaleus coleoptratus dorsal view of male (C) (bar scale: 1,000 μm) and female (GenBank: OL348230 and MZ935645) (D) (bar scale: 1,000 μm) from the blue shark; Pandarus satyrus dorsal view of male (E) (bar scale: 500 μm) and female (GenBank: OL457303 and OL333876) (F) (bar scale: 1,000 μm) from the blue shark; Phyllothyreus cornutus dorsal view of male (G) (bar scale: 1,000 μm) and female (GenBank: MZ935644) (H) (bar scale: 2,000 μm) from the blue shark.
Molecular and Phylogenetic Analyses
High quality sequences for both SSU and cox1 gene loci were successfully obtained for D. latifolia, E. coleoptratus, and P. satyrus. High quality sequences for Ph. cornutus were obtained only for SSU rDNA. The six SSU gene locus sequences obtained from D. latifolia collected from both the shortfin mako shark and the great white shark were identical to each other, and all sequences showed 100% similarity with the sequence (DQ538501) of D. latifolia available in GenBank. The present six cox1 sequences of D. latifolia showed 83–84% similarity with that (KF483702) of Caligus robustus Bassett-Smith, 1898, available in GenBank. Unfortunately, no sequence of D. latifolia for the cox1 gene locus was retrieved from GenBank for comparison.
The sequence of the SSU rDNA obtained from Ph. cornutus showed 100% similarity with the sequence (FJ447449) of Ph. cornutus previously deposited in GenBank. Sequences of E. coleoptratus and P. satyrus were here generated for the first time. The SSU and cox1 sequences obtained for E. coleoptratus showed 98.38 and 86.71% similarity with the sequences of Achtheinus oblongus Wilson, 1908 (FJ447452) and Caligus mutabilis Wilson, 1905 (KF483685) available in GenBank, respectively. The present SSU and cox1 sequences of P. satyrus showed 99.88% and 87% similarity with the sequences of Pandarus sp. 2 (FJ447454-FJ447387) available in GenBank, respectively.
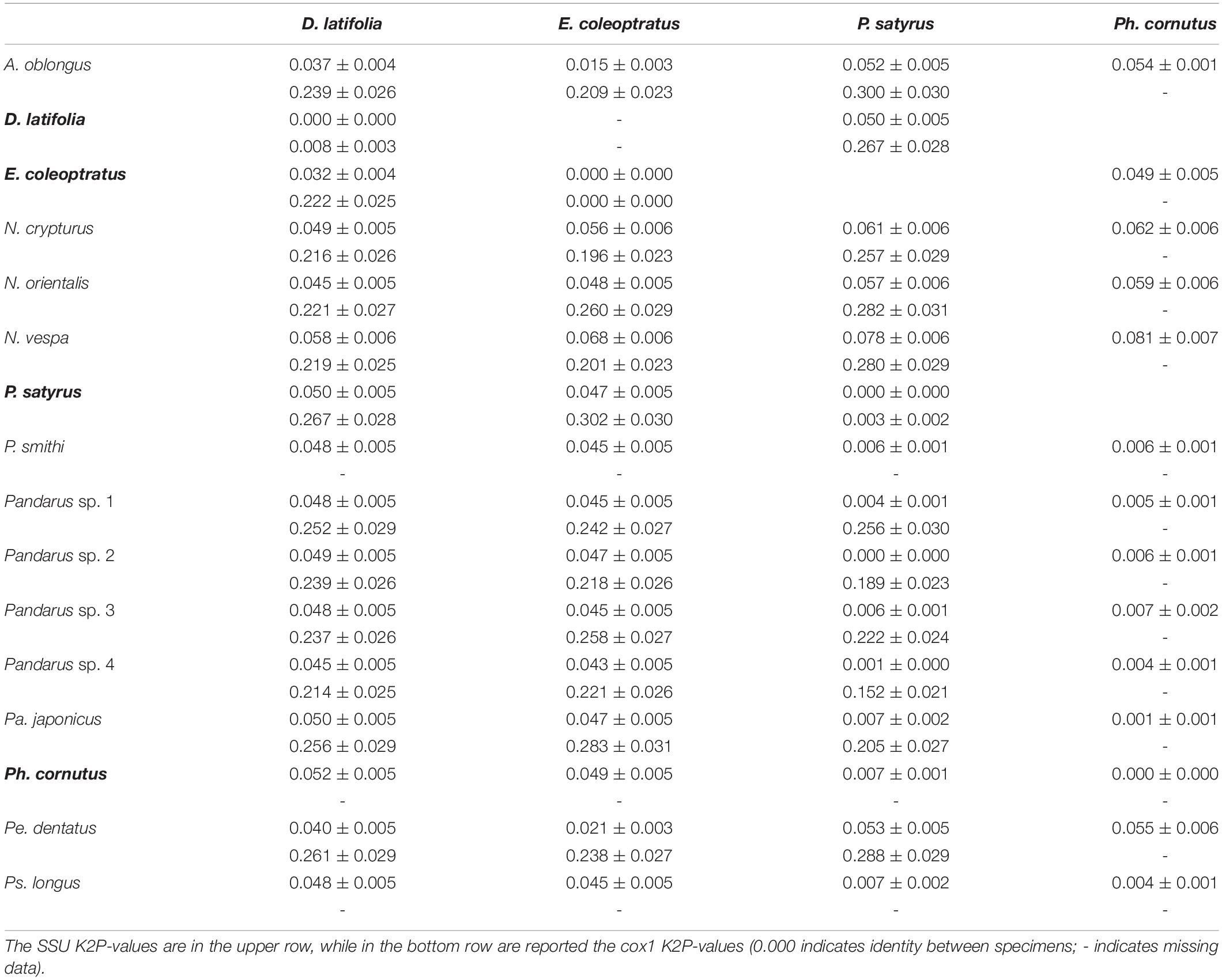
Pairwise distances among specimens and species for the obtained SSU and cox1 sequences are given in Table 3. While no intraspecific variations were found between SSU sequences, intraspecific variations were found in the cox1 sequences of D. latifolia (K2P = 0.008 ± 0.003) and P. satyrus (0.0032 ± 0.002). SSU sequence divergence among species (i.e., interspecific variation) was found to range from a minimum of 0.001 ± 0.000 between P. satyrus and Pandarus sp. 4 to a maximum of 0.081 ± 0.007 between Ph. cornutus and Nesippus vespa Cressey, 1964 (Table 3). Cox1 sequence divergence among species was found to range from a minimum of 0.196 ± 0.023 between E. coleoptratus and Nesippus crypturus Heller, 1865 to a maximum of 0.288 ± 0.029 between P. satyrus and Perissopus dentatus Steenstrup and Lütken, 1861 (Table 3).
Phylogenetic analyses were conducted using both separately (Supplementary Figures 1, 2) and combined cox1 and SSU gene loci (Figure 2). The resulting tree for SSU (Supplementary Figure 1) showed Pandaridae as a monophyletic group, with high support (BS = 100), and the existence of two main clades. The first clade, with strong nodal support (BS = 100) involved two lineages, that included the genera Phyllothyreus Norman, 1903, Pannosus Cressey, 1967, Pseudopandarus Kirtisinghe, 1950 and the paraphyletic genus Pandarus Leach, 1816. At species level, the new generated sequences of P. satyrus clustered with that of Pandarus sp. 2 previously deposited in GenBank in a well-supported lineage (BS = 98). The obtained sequences of Ph. cornutus clustered with high nodal support (BS = 98) with the sequences of Ph. cornutus and Pannosus japonicus (Shiino, 1960) previously deposited in GenBank.
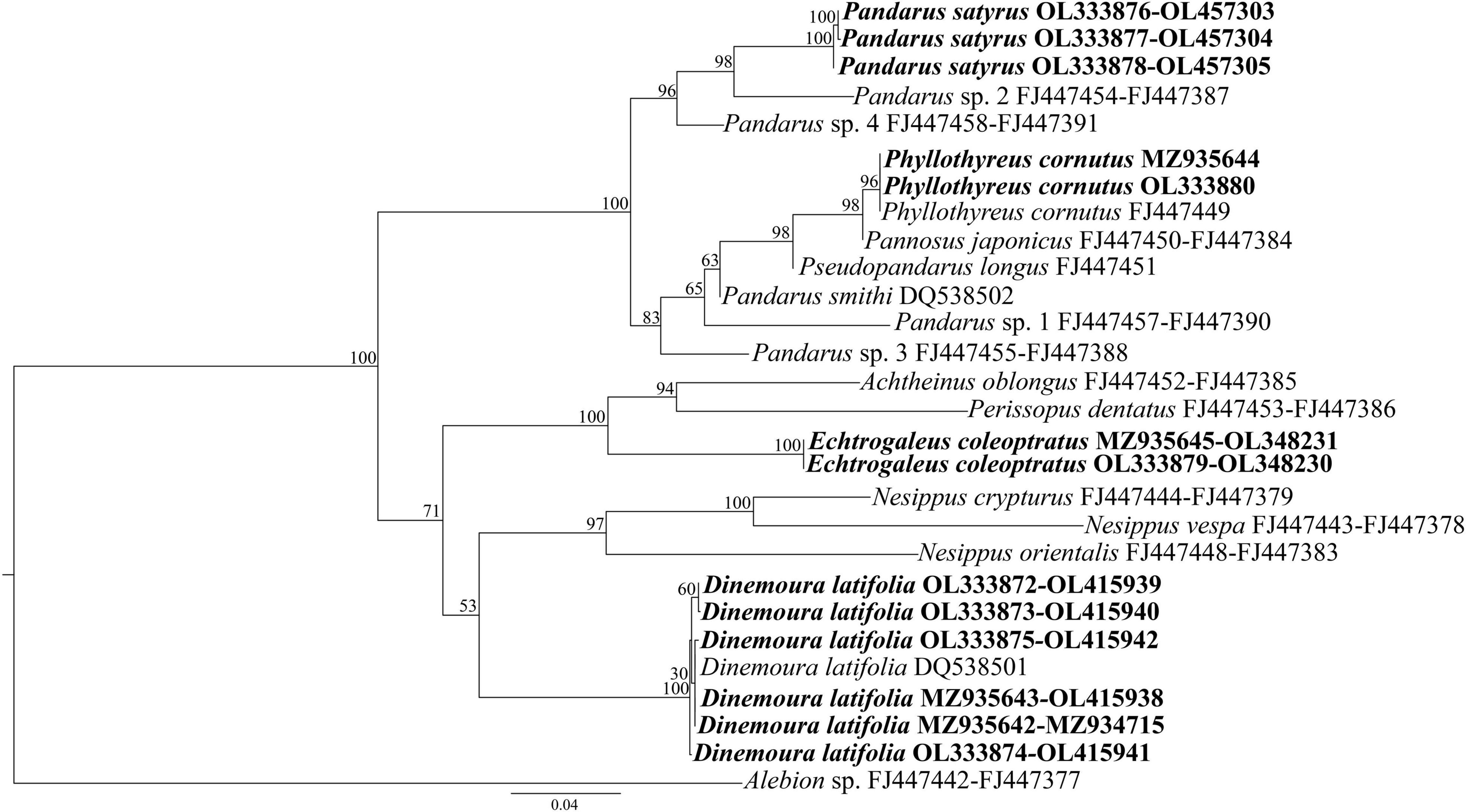
Figure 2. Phylogenetic concatenated tree from maximum likelihood based on SSU and cox1 sequences of pandarid species obtained in the present study, with respect to the pandarid sequences at the same gene loci available in GenBank. Alebion sp. was used as outgroup. The sequences obtained in this study are in bold.
The second major clade, with weaker nodal support (BS = 71), involved three lineages, that included the species of Achtheinus Wilson, 1908, Perissopus Steenstrup and Lütken, 1861, Echtrogaleus Steenstrup and Lütken, 1861, Nesippus Heller, 1865, and Dinemoura Latreille, 1829. At species level, the SSU tree topology placed the new sequences of E. coleoptratus within a well-supported lineage (BS = 99) with the sequences of Achtheinus oblongus and Pe. dentatus, previously deposited in GenBank. The present new generated and the previously deposited sequences of D. latifolia clustered in a separate lineage with high nodal support (BS = 99).
In the resulting tree obtained only for cox1, Pandaridae was also a monophyletic group (BS = 100). Two major clades were generated, the first well-supported (BS = 100) formed by all sequences of Pandarus, Pa. japonicus, A. oblongus, the new generated sequences of E. coleoptratus, N. vespa and Pe. dentatus, and the second (BS = 34) formed by the obtained sequences of D. latifolia and the sequences of N. orientalis and N. crypturus previously deposited in GenBank, highlighting the monophyly of D. latifolia.
The species delimitation analyses of the cox1 gene locus highlighted a total of 10 taxonomic entities, revealing that the sequences of Pandarus spp. belonged to two distinct taxonomic entities (as shown in Supplementary Figure 2). The sequences of P. satyrus obtained in the present study belonged to the same taxonomic entity that included the sequences of Pandarus sp. 2, Pandarus sp. 3, and Pandarus sp. 4 from GenBank (Supplementary Figure 2).
The tree inferred by concatenating the SSU and cox1 gene loci (Figure 2) showed the same topology of the SSU tree (Supplementary Figure 1).
Discussion
This study provides the first molecular data on the occurrence of four species of pandarid copepods from the Mediterranean. To our knowledge prior to of the present study only Ph. cornutus, D. latifolia, E. coleoptratus, and Pandarus bicolor have been recorded on shark species in the Mediterranean Sea (Richiardi, 1880; Brian, 1906; Öktener and Trilles, 2009; Öktener et al., 2020).
In general, pandarid copepods are widely distributed mirroring the movements and distribution of their hosts. In particular, Dinemoura parasitizes the skin of large pelagic sharks. After Cressey (1967), Dinemoura comprises four valid species including D. discrepans Cressey, 1967, D. ferox (Krøyer, 1838), D. latifolia and D. producta (Müller, 1785). The only reports of D. latifolia in the Mediterranean were on blue, shortfin mako, and thresher sharks Alopias vulpinus (Brian, 1906). However, along with its geographical distribution range D. latifolia has been found at least, on other three shark species (i.e., the great white shark, the porbeagle Lamna nasus, and the school shark Galeorhinus galeus) from North and South Atlantic, East, and West Pacific, Indian Ocean, and West Indies (see Williams, 1978).
Phyllothyreus cornutus, the only species in its monotypic genus, infects the gills of several pelagic sharks (i.e., the blue shark, the shortfin mako shark, the porbeagle, the smooth hammerhead Sphyrna zygaena, the sandbar shark Carcharhinus plumbeus, and the tiger shark Galeocerdo cuvier) from the North and South Atlantic and North Pacific (Hewitt, 1967; Schaeffner and Smit, 2019). In the Mediterranean Sea, it has been reported exclusively on the blue shark (Richiardi, 1880).
The genus Echthrogaleus comprises eight species including E. asiaticus Ho, Liu and Lin, 2012, E. coleoptratus, E. denticulatus Smith, 1873, E. disciarai Benz and Deets, 1987, E. mitsukurinae Izawa, 2012, E. pellucidus Shiino, 1963, E. spinulus Morales-Serna, Crow, Montes and González, 2019 and E. torpedinis Wilson, 1907. Echthrogaleus coleoptratus has been reported from the North and South Atlantic, the Pacific and Indian Oceans, and the Mediterranean Sea (Hewitt, 1967). It parasitizes the skin of about 13 species of sharks; however, it is commonly found on the great white shark, the porbeagle and the blue shark (Hewitt, 1967, 1979; Cressey and Lachner, 1970; Rokicki and Bychawska, 1991; Henderson et al., 2002; Benz et al., 2003; Luque and Tavares, 2007). In the Mediterranean, it has been reported on the blue shark, the gulper shark Centrophorus granulosus (Bloch and Schneider, 1801) and the great white shark (Brian, 1906).
The genus Pandarus comprises 14 nominal species including P. ambiguous (Scott, 1907), P. bicolor, P. brevicaudis Dana, 1852, P. carcharhini Ho, 1963, P. cranchii Leach, 1819, P. floridanus Cressey, 1967, P. katoi Cressey, 1967, P. niger Kirtisinghe, 1950, P. rhincodonicus Norman, Newbound and Knott, 2000, P. rouxii Risso, 1826, P. satyrus Dana, 1849, P. sinuatus Say, 1818, P. smithii and P. zygaenae Brady, 1883. Pandarus satyrus has a wide geographical distribution including Atlantic, Pacific, and Indian Ocean; however, it has never been reported from the Mediterranean. According to Cressey (1967); Benz (1986), and Rojas et al. (2001), P. satyrus has been predominantly found on the blue shark. It is closely related to P. cranchii with which it was synonymized by Shiino (1954) but considered as valid species by Cressey (1967). According to Cressey (1967) the two species are easily separated on the basis of the caudal rami. The rami of P. cranchii extend at least to the tip of the abdominal plate (often beyond) whereas the rami of P. satyrus extends only about half the length of the abdominal plate. The only other species of Pandarus reported from the Mediterranean is P. bicolor found on the blue shark, the dusky smooth-hound, the common smooth-hound, the angular rough shark Oxynotus centrina Linnaeus, 1758 and the picked dogfish Squalus acanthias Linnaeus, 1758 (Richiardi, 1880; Brian, 1906; Öktener and Trilles, 2009; Öktener et al., 2020). Pandarus bicolor can be distinguished from P. satyrus as the cephalon only occupies 1/3 of the total body length and its caudal rami are small and scarcely visibly dorsally (Cressey, 1967).
Based on specific morphological characters, pandarid copepods have been arranged into two major groups: (i) species with all three thoracic segments provided with dorsal or dorsolateral plates (Pandarus-group), and (ii) species with the second free thoracic segment without plates (Dinemoura-group) (Kabata, 1979). Pandarus and Phyllothyreus have been included in the first group with Achtheinus, Perissopus, Gangliopus Gerstaecker, 1854, Pannosus and Pseudopandarus; Dinemoura and Echtrogaleus have been included in the second group with Demoleus Heller, 1865, Dinemoleus Cressey and Boyle, 1978, Nesippus, Paranesippus Shiino, 1955 and Pagina Cressey, 1963 (see Kabata, 1979).
In contrast, based on the results of more recent phylogenetic analyses, pandarid copepods have been placed into two major clades: the first clade included the species of Nesippus, and the second clade included the species of Phyllothyreus, Pannosus, Pandarus, Pseudopandarus, and Achtheinus (Dippenaar, 2009). However, Dippenaar (2009) focused mainly on relationships among families of the Siphonostomatoida, while no phylogenetic relationships were deepened among the genera.
Maximum-likelihood analysis inferred by concatenated SSU + cox1 data set placed the sequences of pandarid copepods available in GenBank plus the new generated sequences into two major clades, however, some differences were observed when the present results were compared to those of Dippenaar (2009). For example, the present specimens of P. satyrus and Ph. cornutus were included in a first clade with Pandarus spp., Pa. japonicus, Ps. longus, and P. smithi with strong nodal support. Within this clade, Ph. cornutus, Pa. japonicus, Ps. longus, P. smithi, Pandarus sp. 1, and Pandarus sp. 3 were placed in a subclade not supported by the posterior probabilities and bootstrap analysis. Finally, D. latifolia and E. coleoptratus were included in a second clade with a weaker nodal support with Nesippus orientalis Heller 1865, N. vespa, N. crypturus, A. oblongus, and Pe. dentatus.
The phylogenetic pattern for the species here collected was congruent with the morphological characters of the two species groups above mentioned, except for A. oblongus and Pe. dentatus which were placed into the second major clade with the genera Echtrogaleus, Nesippus and Dinemoura. In contrast, the phylogenetic clustering among the members of the two clades seems to be not related to the host preference. For instance, both clades included parasites capable of infecting shark species belonging to six orders and 11 families, with the second clade that also included parasites capable of infecting five additional families of sharks. Nevertheless, we cannot exclude a coevolutionary hypothesis between copepods and their hosts species. Indeed, little is known regarding the nature of host-copepod association in elasmobranchs (Bernot et al., 2021). Huys et al. (2007) suggested a host switching event in the siphonostomatoid copepods highlighting monostrilloids’ alterations in the host utilization, body plan, and life cycle strategy. The scarcity of data regarding the host association and life cycle strategy of pandarid copepods does not help to resolve phylogenetic relationships among species. The present phylogenetic analysis included only a small subset (12) of the 64 valid species of Pandaridae. Therefore, it is possible that the present phylogenetic results may not reflect the true relationships, as a large majority of species is missing from the present analysis. Indeed, as already discussed above, our phylogenetic results were not congruent with those obtained by Dippenar (2009). More thorough sequencing of Pandaridae species will be needed to better resolve the phylogenetic relationships among the members of this family.
Conclusion
In conclusion, we provide additional DNA sequences for D. latifolia and Ph. cornutus. Furthermore, new molecular data for E. coleoptratus and P. satyrus are reported, based on, both the nuclear (SSU) and mitochondrial (cox1) gene loci. Whilst the single use of the SSU gene locus permitted the molecular identification of the copepod species, the mtDNA cox1 could represent a suitable marker to infer population structure of pandarid copepods, and consequently of their hosts (Criscione et al., 2006; Baldwin et al., 2011). In this sense, intraspecific variation of cox1 was actually detected for some of the species in the present study. Nonetheless, the scarce reference sequence information, hampered any further understanding on the population structure of these copepod parasites. This study represents the first attempt to yield new molecular and phylogenetic data of pandarid copepods on pelagic sharks in the Mediterranean Sea that could contribute to a better characterization of these poorly known parasites. Future molecular and genetic studies should also provide a more detailed assessment of the host-parasite interactions, ecological data, and life cycle strategy. Pandarus satyrus represents a new record for the Mediterranean.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI accession numbers: MZ935642-45, OL333872-80, MZ934715, OL415938-42, OL348230-31, and OL457303-05.
Ethics Statement
The material here studied has been collected from two co-authors (GI and BZ) of the present paper under the framework of a project of the Museo Civico di Storia Naturale (MSNC) in Comiso on alien and rare marine species of the Mediterranean Sea. The MSNC is a scientific institution registered at the CITES Secretariat, D.M. 23.03.1994 (Cod. IT030), authorized to take, keep, use and display dead endangered species of wild fauna.
Author Contributions
MP and MS design of experiment, performed morphological and molecular analyses, and wrote the manuscript. GI and BZ collected the host data and parasitic copepods. All authors approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank all people involved with shark carcass recovery. In particular, we wish to thank the scuba divers Giovanni Pagano and Tonino Giunta (Pozzallo), the fishermen Salvatore Avarino (Marzamemi) and Ignazio Conti (Donnalucata), Domenico Mirabella of the vessel “Trinacria” (Catania) and Marta Nicosì (Marina di Ragusa). We thank the port authorities of the Sicily Region, the Coast Guard and the Cites Service of the Nucleo Carabinieri, who have contributed to the recovery of the shark individuals stranded or bycatch.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.778034/full#supplementary-material
Supplementary Figure 1 | Phylogenetic tree from maximum likelihood based on SSU sequences of pandarid species obtained in the present study, with respect to the pandarid sequences at the same gene locus available in GenBank. Alebion sp. was used as outgroup. The sequences obtained in this study are in bold.
Supplementary Figure 2 | Phylogenetic tree from maximum likelihood based on cox1 sequences of pandarid species obtained in the present study, with respect to the pandarid sequences at the same gene locus available in GenBank. Alebion sp. was used as outgroup. The sequences obtained in this study are in bold. Braces indicate the specimens that according to the species delimitation analyses are part of the same taxonomic entity. Numbers in brackets indicate the 10 taxonomic entities revealed by the species delimitation analyses.
References
Akaike, H. (1973). “Information theory and an extension of the maximum likelihood principle,” in Proceedings of the Second International Symposium on Information Theory, eds B. N. Petrov and F. Caski (Budapest: Akademiai Kiado), 267–281.
Alvarez, F., and Winfield, I. (2001). New records of Dinemoura latifolia and Pandarus smithii (Copepoda, Siphonostomatoida, Pandaridae) parasitizing the shark Isurus oxyrinchus in the Gulf of Mexico. Crustaceana 74, 501–503. doi: 10.1163/156854001750243072
Baldwin, R. E., Beth Rew, M., Johansson, M. L., Banks, M. A., and Jacobson, K. C. (2011). Population Structure of three species of Anisakis nematodes recovered from Pacific sardines (Sardinops sagax) distributed throughout the California current system. J. Parasitol. 97, 545–554. doi: 10.1645/GE-2690.1
Benz, G. W. (1981). Observations on the attachment scheme of the parasitic copepod Pandarus satyrus (Copepoda: Pandaridae). J. Parasitol. 67, 966–967. doi: 10.2307/3280732
Benz, G. W. (1986). Distribution of siphonostomatoid copepods parasitic upon large pelagic sharks in the western North Atlantic. Syllogeus 58, 211–219.
Benz, G. W., Mollet, H. F., Ebert, D. A., Davis, C. R., and Van Sommeran, S. R. (2003). Five species of parasitic copepods (Siphonostomatoida: Pandaridae) from the body surface of a white shark captured in Morro Bay, California. Pac. Sci. 57, 39–43. doi: 10.1353/psc.2003.0002
Bernot, J. P., and Boxshall, G. A. (2017). A new species of Pseudopandarus Kirtisinghe, 1950 (Copepoda: Siphonostomatoida: Pandaridae) from sharks of the genus Squalus L. in New Caledonian waters. Syst. Parasitol. 94, 275–291. doi: 10.1007/s11230-016-9692-2
Bernot, J. P., Boxshall, G. A., and Crandall, K. A. (2021). A synthesis tree of the Copepoda: integrating phylogenetic and taxonomic data reveals multiple origins of parasitism. PeerJ. 9:e12034. doi: 10.7717/peerj.12034
Compagno, L. J. V. (1984). Sharks of the world. An annotated and illustrated catalogue of shark species known to date. Hexanchiformes to Lamniformes. Rome: Food and Agriculture Organization of the United Nations.
Cressey, R. F. (1967). Revision of the family Pandaridae (Copepoda: Caligoida). Proc. USA Natl. Mus. 121, 1–133. doi: 10.5479/si.00963801.121-3570.1
Cressey, R. F. (1968). Caligoid copepods parasitic on Isurus oxyrinchus with an example of habitat shift. Proc. USA Natl. Mus. 125, 1–26. doi: 10.5479/si.00963801.125-3653.1
Cressey, R. F., and Lachner, E. A. (1970). The parasitic copepod diet and life history of diskfishes (Echeneidae). Copeia 1970, 310–318. doi: 10.2307/1441652
Criscione, C. D., Cooper, B., and Blouin, M. S. (2006). Parasite genotypes identify source populations of migratory fish more accurately than fish genotypes. Ecology 87, 823–828. doi: 10.1890/0012-9658(2006)87[823:PGISPO]2.0.CO;2
Deidun, A., Insacco, G., Galdies, J., Balistreri, P., and Zava, B. (2021). Tapping into hard-to-get information: the contribution of citizen science campaigns for updating knowledge on range-expanding, introduced and rare native marine species in the Malta-Sicily Channel. Bioinvasions Rec. 10, 257–269. doi: 10.3391/bir.2021.10.2.03
Dippenaar, S. M. (2009). Estimated molecular phylogenetic relationships of six siphonostomatoid families (Copepoda) symbiotic on elasmobranchs. Crustaceana 82, 1547–1567. doi: 10.1163/001121609X12511103974538
Folmer, O., Black, M., Hoeh, W., Lutz, R., and Vrijenhoek, R. (1994). DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 3, 294–299.
Gunn, A., and Pitt, S. J. (2012). Parasitology: An Integrated Approach. Oxford: Wiley-Blackwell. doi: 10.1002/9781119968986
Henderson, A. C., Flannery, K., and Dunne, J. (2002). An investigation into the metazoan parasites of the spiny dogfish (Squalus acanthias L.), off the west coast of Ireland. J. Nat. Hist. 36, 1747–1760. doi: 10.1080/00222930110066431
Hewitt, G. C. (1967). Some New Zealand parasitic Copepoda of the family Pandaridae. NZJ Mar. Freshw. Res. 1, 180–264. doi: 10.1080/00288330.1967.9515202
Hewitt, G. C. (1979). Eight species of parasitic Copepoda on a white shark. NZJ Mar. Freshw. Res. 13:171. doi: 10.1080/00288330.1979.9515790
Huys, R., Llewellyn-Hughes, J., Conroy-Dalton, S., Olson, P. D., Spinks, J. N., and Johnston, D. A. (2007). Extraordinary host switching in siphonostomatoid copepods and the demise of the Monstrilloida: integrating molecular data, ontogeny and antennulary morphology. Mol. Phylogenet. Evol. 43, 368–378. doi: 10.1016/j.ympev.2007.02.004
Izawa, K. (2010). Free-living stages of the parasitic copepod, Gangliopus pyriformis Gerstaecker, 1854 (Siphonostomatoida, Pandaridae) reared from eggs. Crustaceana 83, 829–837. doi: 10.1163/001121610X498863
Kabata, Z. (1979). “Parasitic Copepoda of the British fishes,” in Crustaceana, ed. J. H. Stock (London: The Bristish Museum), 468.
Kabata, Z. (1988). “Copepoda and Branchiura,” in Guide to the Parasites of Fishes of Canada Part II – Crustacea, eds L. Margolis and Z. Kabata (Ottawa, ON: Publié par), 3–127.
Katsanevakis, S., Poursanidis, D., Hoffman, R., Rizgalla, J., Rothman, S. B. S., Levitt-Barmats, Y., et al. (2020). Unpublished Mediterranean records of marine alien and cryptogenic species. Bioinvasions Rec. 9, 165–182. doi: 10.3391/bir.2020.9.2.01
Kimura, M. (1980). A simple model for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120. doi: 10.1007/BF01731581
Kumar, S., Stecher, G., Michael, L., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Lanfear, R., Calcott, B., Ho, S. Y. W., and Guindon, S. (2012). Partitionfinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol. Biol. Evol. 29, 1695–1701. doi: 10.1093/molbev/mss020
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Lewis, A. G. (1966). Copepod crustaceans parasitic on elasmobranch fishes of the Hawaiian Islands. Proc. USA Natl. Mus. 118, 57–154. doi: 10.5479/si.00963801.118-3524.57
Luque, J. L., and Tavares, L. R. (2007). Checklist of Copepoda associated with fishes from Brazil. Zootaxa 1579, 1–39. doi: 10.11646/zootaxa.1579.1.1
Morgulis, A., Coulouris, G., Raytselis, Y., Madden, T. L., Agarwala, R., and Schäffer, A. A. (2008). Database indexing for production MegaBLAST searches. Bioinformatics 24, 1757–1764. doi: 10.1093/bioinformatics/btn322
Nguyen, L. T., Schmidt, H. A., von Haeseler, A., and Minh, B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Öktener, A., and Trilles, J. P. (2009). Four parasitic copepods on marine fish (Teleostei and Chondrichthyes) from Turkey. Acta Adriat. 50, 121–128.
Öktener, A., Ventura, D., and Şirim, M. (2020). Occurrence of Pandarus bicolor (Siphonostomatoida:Pandaridae) on vulnerable shark species: Oxynotus centrina and Sqaulus acanthias from Turkish Marine Waters. Vie. Milieu 70, 19–32.
Puillandre, N., Brouillet, S., and Achaz, G. (2020). ASAP: assemble species by automatic partitioning. Mol. Ecol. Resour. 21, 609–620. doi: 10.1111/1755-0998.13281
Richiardi, S. (1880). “Catalogo sistematico dei Crostacei che vivono sul corpo degli animali acquatici in Italia,” in Catalogo Generale Della Sezione Italiana Alla Esposizione Internazionale Della Pesca in Berlino, eds T. C. Walter and G. Boxshall (Berlino: Internationalen Fischerei Ausstellung), 146–153.
Rojas, M. J. R., Rodríguez Solano, O., and Morales-Ramírez, A. (2001). Size and distribution of Pandarus satyrus (Copepoda: Pandaridae) on the blue shark Prionace glauca (Carcharhiniformes: Carcharhinidae) in Costa Rica. Rev. Biol. Trop. 49, 199–201.
Rokicki, J., and Bychawska, D. (1991). Parasitic copepods of Carcharhinidae and Sphyridae (Elasmobranchia) from the Atlantic Ocean. J. Nat. Hist. Mus. 25, 1439–1448. doi: 10.1080/00222939100770911
Santoro, M., Iaccarino, D., and Bellisario, B. (2020). Host biological factors and geographic locality influence predictors of parasite communities in sympatric sparid fishes off the southern Italian coast. Sci. Rep. 10:13283. doi: 10.1038/s41598-020-69628-1
Santoro, M., Mattiucci, S., Cipriani, P., Bellisario, B., Romanelli, F., Cimmaruta, R., et al. (2014). Parasite communities of icefish (Chionodraco hamatus) in the Ross Sea (Antarctica): Influence of the host sex on the helminth infracommunity structure. PLoS One 9:e88876. doi: 10.1371/journal.pone.0088876
Schaeffner, B. C., and Smit, N. J. (2019). Parasites of cartilaginous fishes (Chondrichthyes) in South Africa - a neglected field of marine science. Folia Parasitol. 66:2. doi: 10.14411/fp.2019.002
Shiino, S. M. (1954). Copepods parasitic on Japanese fishes. 5: five species of the family Pandaridae. Rep. Fac. Fish. Pref. Univ. Mie 1, 291–332.
Vaidya, G., Lohman, D. J., and Meier, R. (2011). SequenceMatrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 27, 171–180. doi: 10.1111/j.1096-0031.2010.00329.x
Walter, T. C., and Boxshall, G. (2021). World of Copepods Database. Pandaridae Milne Edwards, 1840. Available online at: http://www.marinespecies.org/aphia.php?p=taxdetails&id=135531 (accessed June 21, 2021).
Williams, E. H. (1978). Conchoderma virgatum (Spengler) (Cirripedia Thoracica) in association with Dinemoura latifolia (Steenstrup & Lutken) (Copepoda, Caligidea), a parasite of the shortfin mako, Isurus oxyrhynchus Rafinesque (Pisces, Chondrichthyes). Crustaceana 34, 109–110. doi: 10.1163/156854078X00655
Keywords: parasitic copepods, Pandaridae, Carcharodon carcharias, Isurus oxyrinchus, Prionace glauca, molecular identification, phylogenetic analysis
Citation: Palomba M, Insacco G, Zava B and Santoro M (2022) Occurrence and Molecular Characterization of Some Parasitic Copepods (Siphonostomatoida: Pandaridae) on Pelagic Sharks in the Mediterranean Sea. Front. Mar. Sci. 8:778034. doi: 10.3389/fmars.2021.778034
Received: 16 September 2021; Accepted: 23 December 2021;
Published: 28 January 2022.
Edited by:
Nathan Jack Robinson, Fundación Oceanográfica, SpainCopyright © 2022 Palomba, Insacco, Zava and Santoro. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mario Santoro, mario.santoro@szn.it
 Marialetizia Palomba
Marialetizia Palomba Gianni Insacco2
Gianni Insacco2  Mario Santoro
Mario Santoro